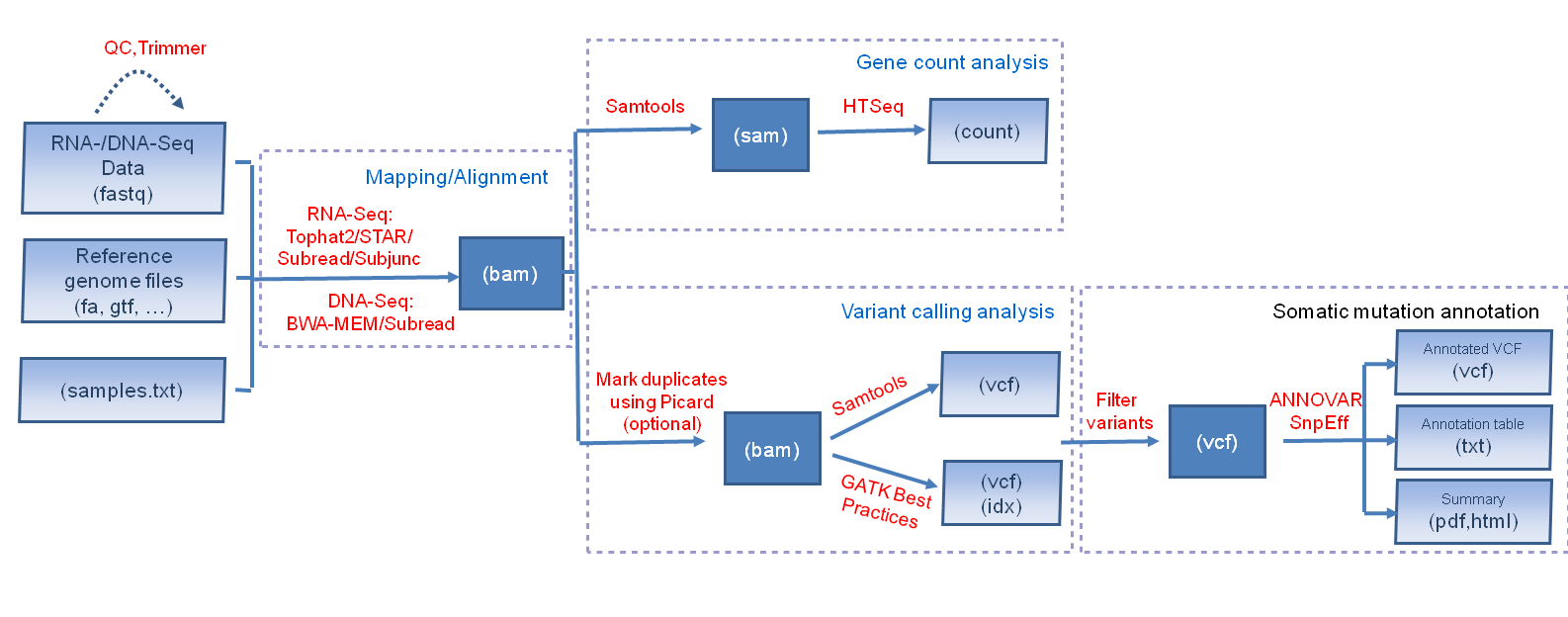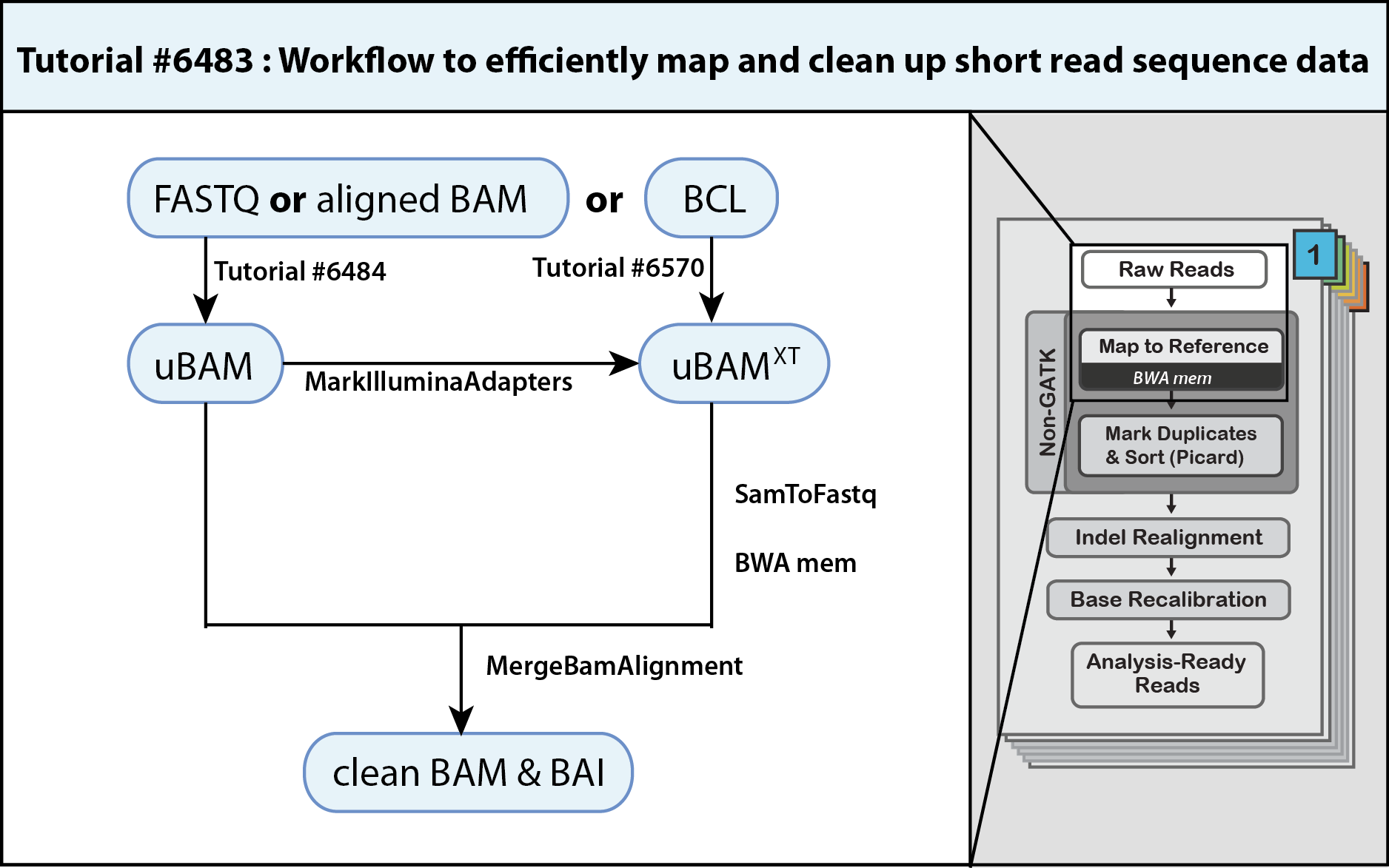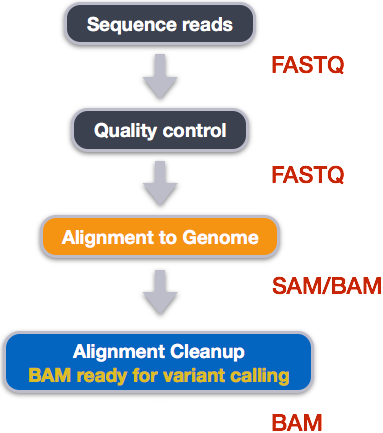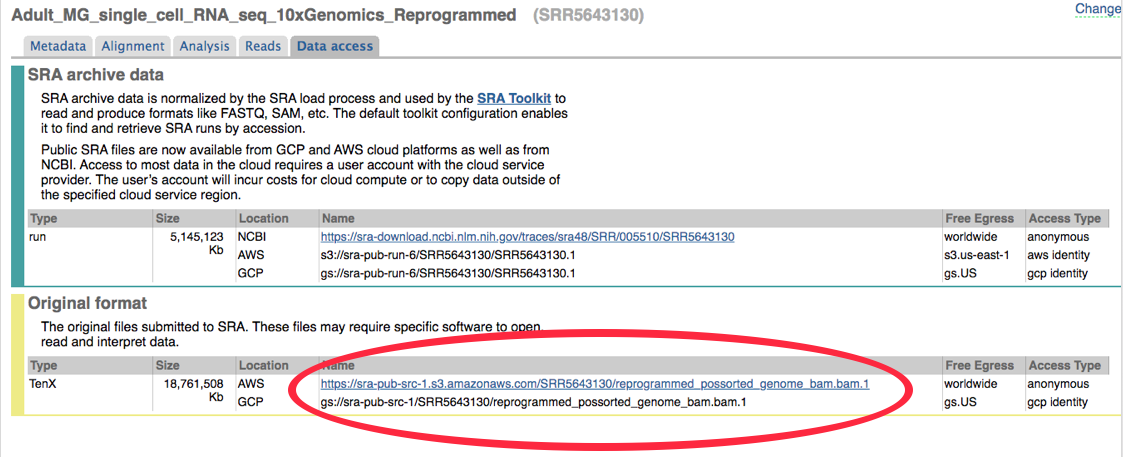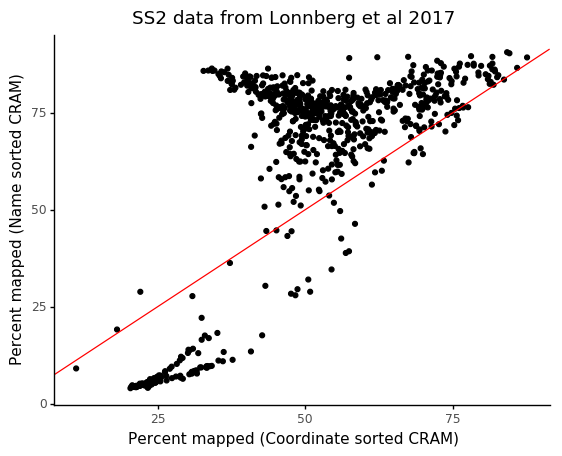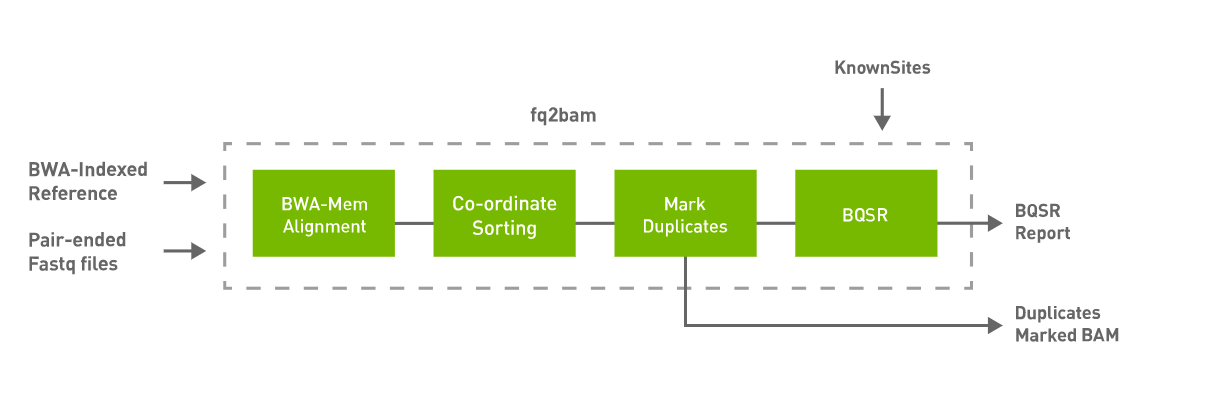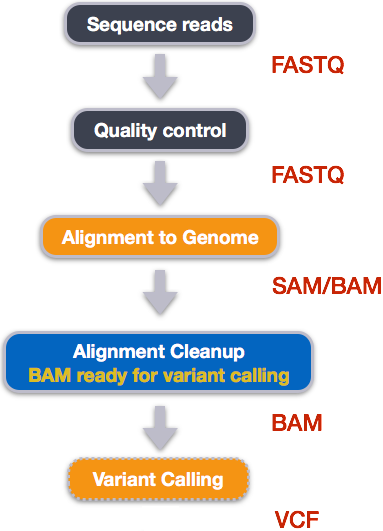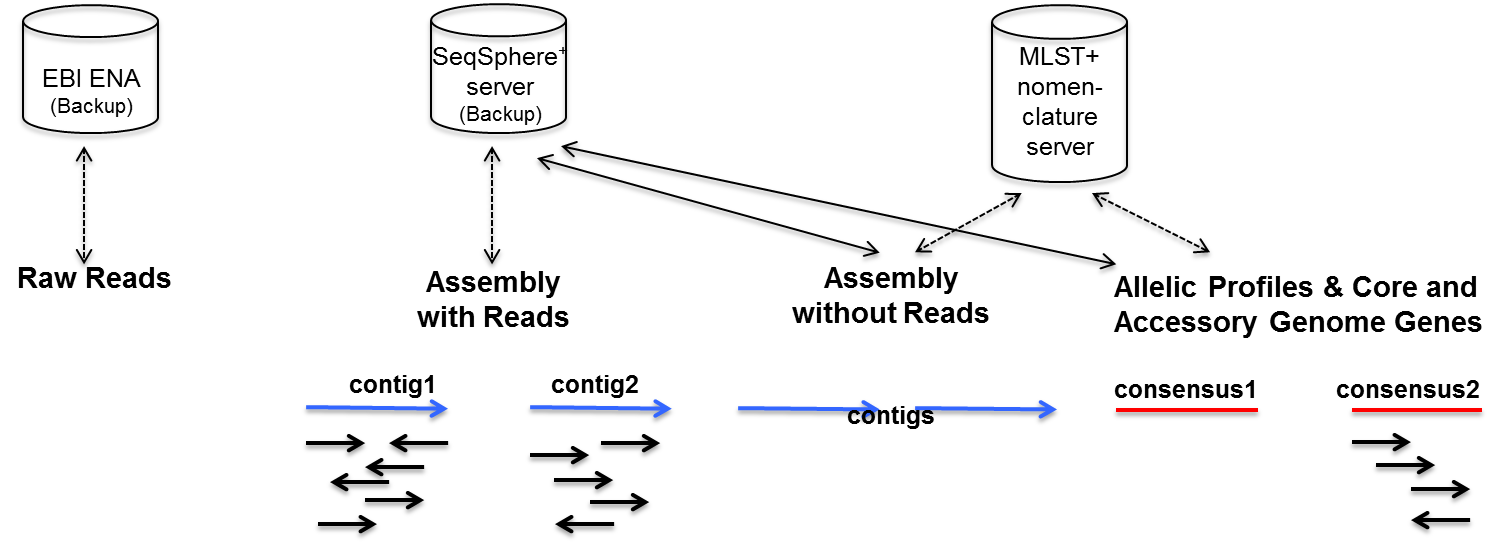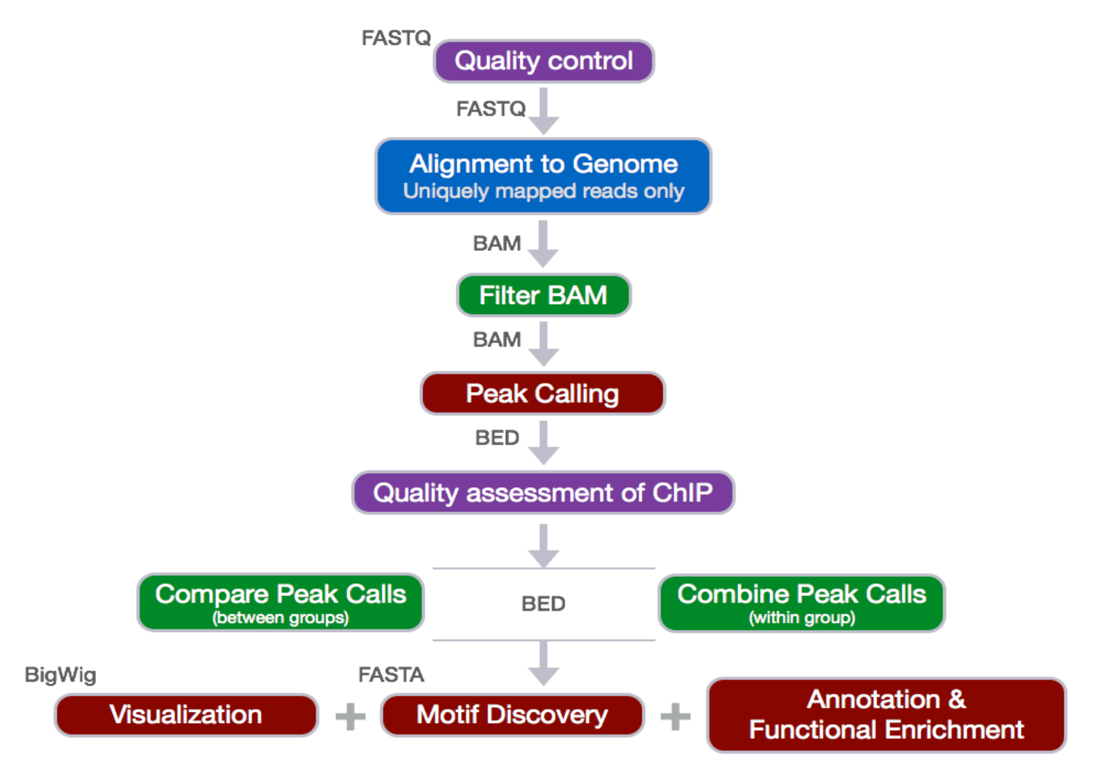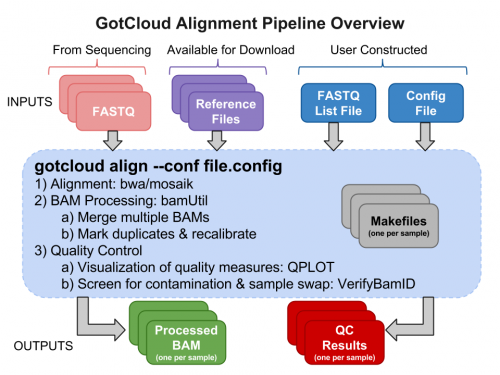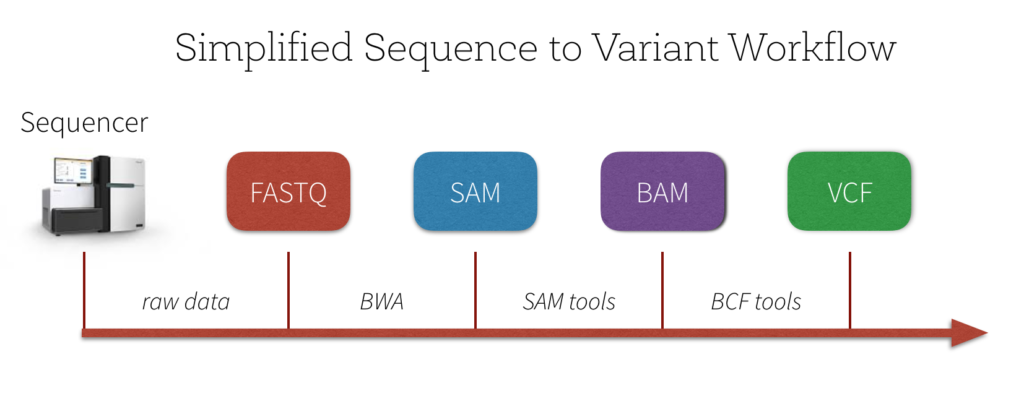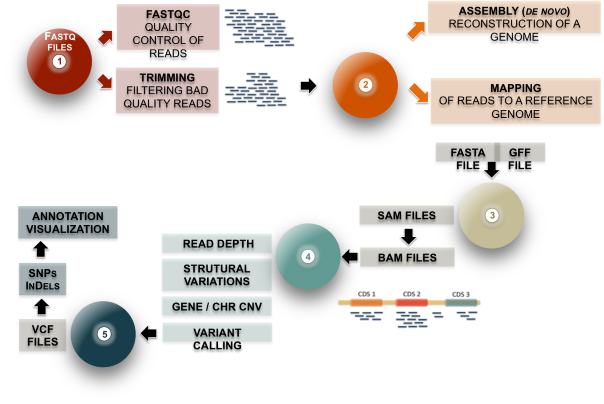
3. Day 2: Introduction to basic NGS pipelines — CODATA-RDA Research Data Science Advanced Workshop on Bioinformatics 1.0 documentation
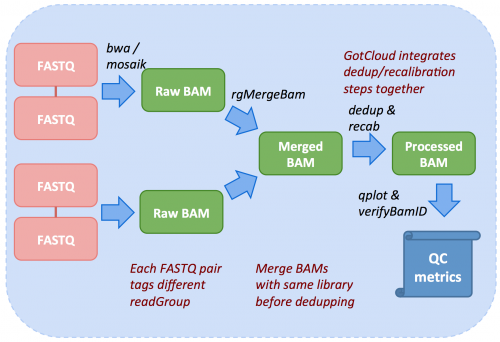
gatk/doc_archive/deprecated/[How_to]_Generate_a_BAM_for_variant_discovery_(long).md at master · broadgsa/gatk · GitHub